It lists one mutation per row, and the columns named in the header row report several pieces of information for each mutation. The columns of this file are: The mutational spectrum model does not consider every possible base change on its own, but pools each mutation into categories that consider both sequence context e. Indelocator How do I get Indelocator? After a free registration, you can access it here:. 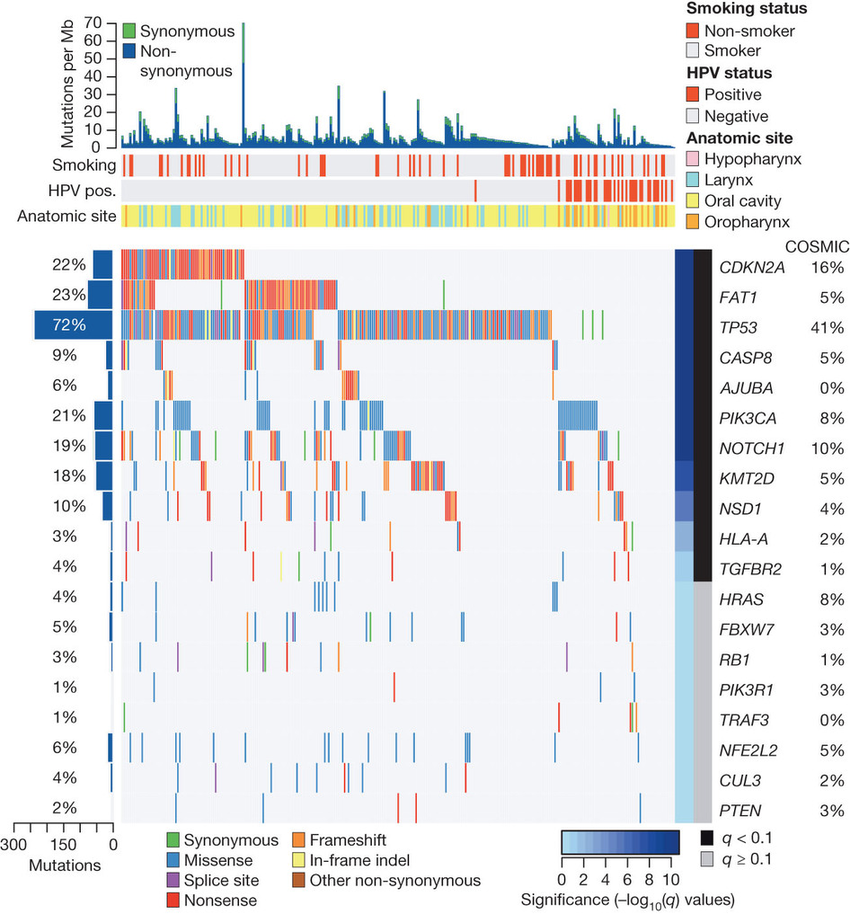
| Uploader: | Jutaur |
| Date Added: | 9 April 2015 |
| File Size: | 68.80 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 13956 |
| Price: | Free* [*Free Regsitration Required] |
For the waterfall plot there is a nice tutorial here: However, it appears that one difficulty with larger sample size is that it increases the rate of detecting highly mutable genes as not simply highly mutable, but also implausibly cancer-related -- that is, it increases the false positive rate. In a mock case where the genes were given variable mutation frequencies and the dataset was analyzed under the erroneous assumption of a constant genome-wide mutation rate, many of the highly mutable genes were falsely detected as significant.
Genes in closed chromatin as measured by HiC or ChipSeq have higher mutation rates than genes in open chromatin. In particular, the dynamic range of MutSigCV1.
Each mutation belongs to mhtsigcv one category. For more information on the file contents and format, see the Input Files section.
Mutational heterogeneity in cancer and the search for new cancer-associated genes.
Nature Algorithm A critical component of MutSigCV is the background model for mutations, the probability that a base is mutated by chance. I am trying to run MutSig with with the three minimal input files and output path: Recent cancer genome studies have led mutssigcv the identification of many cancer genes, and the expectation was that as samples sizes grew, the power to detect cancer driver genes sensitivity and distinguish them from the background of random mutsicgv specificity would increase as well.
How do I run ContEst? How do I prepare the input files? If it is a null mutation or indel, it belongs in the last category.
MutSig |
A tab-delimited file that gives the maximum number of bases covered to adequate depth in order to call mutations. It is also broken down by "categ" and "effect" as listed above. How do I get help? How do I run MuTect? These neighbor genes are chosen on the basis of having similar genomic properties to the central gene in question: The "nnei","x", and "X" values in the MutSig output analysis give insight into how the background mutation rate is calculated for a given gene.
How do I get MutSig?
Then this tally is converted to a score, and then to a mutxigcv level. How do I run ContEst? Note that though MutSigCV was developed for the analysis of somatic mutations, it has also been used successfully with germline mutations. The Mutation Annotation Format MAF file is a tab-delimited text file that contains information about the mutations detected in the sequencing project. These studies involve the sequencing of matched tumour-normal samples followed by mathematical analysis to identify those genes in which mutations occur more frequently than expected by random chance.
Hi all, I am working on DNA data our own experimental data for some cancer types of human. Within each gene-patient bin, the coverage is broken down further according to the mutation category e. In such cases, a reasonable approach is to carry out the computation assuming full coverage. Major international projects are underway that are aimed at creating a comprehensive catalogue of all the genes responsible for the initiation and progression of cancer.
Please mutsgicv the authors listed above if you have an interest in installing MutSigCV locally.
How do I run MutSig? |
The four columns required by the program are: Other options listed here. Email Required, but mtsigcv shown. MutSigCV starts from the observation that the data is very sparse, and that mutslgcv are usually too few silent mutations in a gene for its BMR to be estimated with any confidence.
The mutations from all the tumors can be aggregated together by collapsing as shown, and the total number of mutations per gene can be computed. Be sure to replace mutations.


Комментариев нет:
Отправить комментарий